..
IMAGES FROM CHAPTER 9:
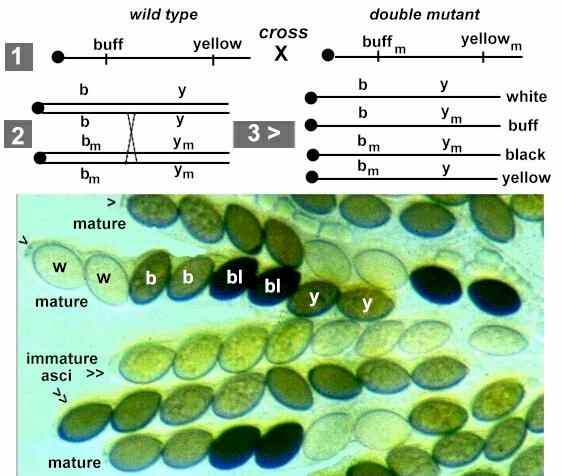
Fig. 9.1. Illustration of the segregation of spore colour genes during the first division (A) or second division (B) of meiosis in Ascomycota. See text for details. [© Jim Deacon]
Fig. 9.2. Part of a crushed perithecium of Sordaria, showing several asci containing 8 ascospores. In the normal, intact perithecium the ascospores would be released through small pores (arrowheads) at the ascus tips. [Courtesy of C. Charier & D.J.Bond, The University of Edinburgh]
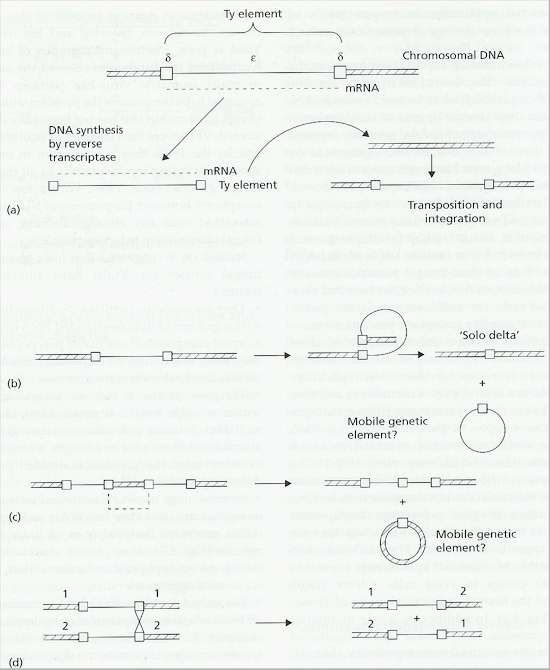
Fig. 9.3. Ty elements of yeast. A: The Ty element consists of an epsilon region which encodes its own replication, flanked by delta sequences. Copies of the transposon are then inserted elsewhere in the same or in other chromosomes. B: The delta regions of one Ty element can undergo homologous recombination, leading to a ‘solo delta’ in the chromosome. C: Homologous recombination between two Ty elements on a chromosome could lead to excision of part of the chromosome. D: Homologous recombination between Ty elements on different chromosomes could create hybrid chromosomes. [Based on a diagram in S.G.Oliver, 1987, pp. 33-52 in: Evolutionary Biology of the Fungi, (eds A.D.M.Rayner, C.M.Brasier & D.Moore) Cambridge University Press]
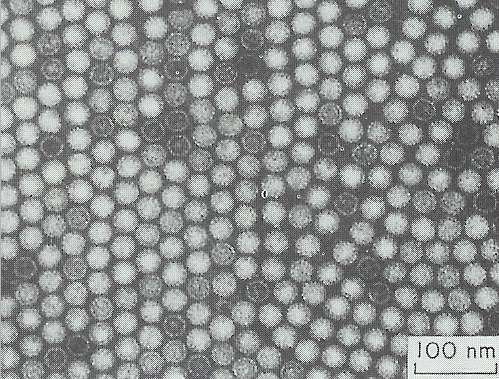
Fig. 9.4. Isometric virus-like particles extracted from hyphae of Colletotrichum sp. The particles aggregate in crystalline arrays in vitro. [Courtesy of C.J.Rawlinson et al. (1975) Transactions of the British Mycological Society 65, 305-308]
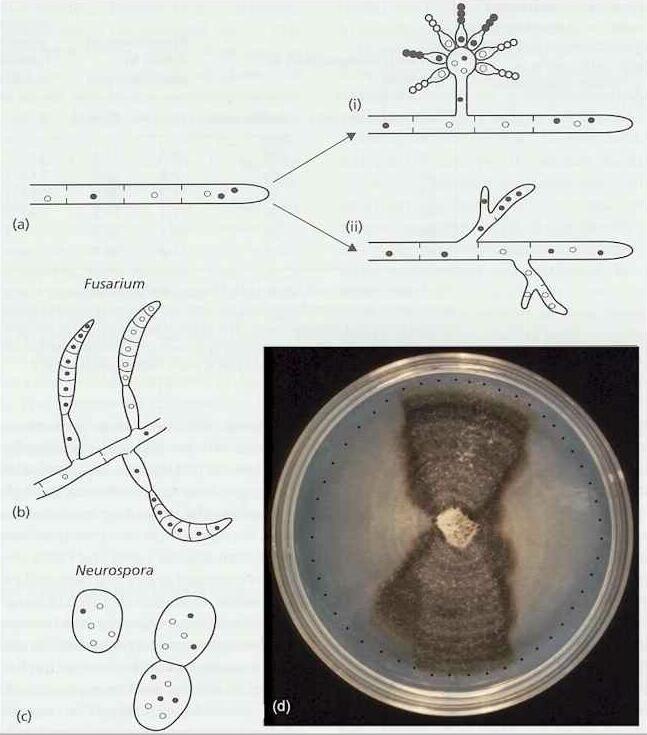
Fig. 9.5. Heterokaryosis and the reversion to homokaryons. See text for details. [© Jim Deacon]
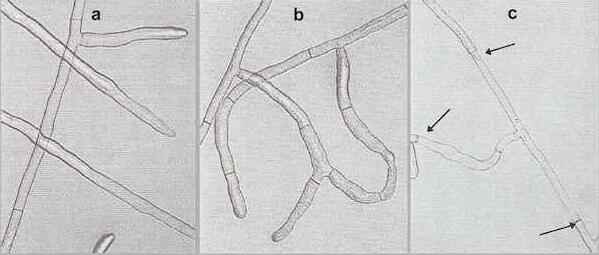
Fig. 9.6. Anastomosis reactions of Rhizoctonia solani. (a) No reaction, when strains of different anastomosis groups show no hyphal attraction. (b) Compatible reaction, when strains of the same anastomosis group orientate towards one another and fuse to form a continuous hyphal network. (c) Incompatible reaction, when strains of the same anastomosis group but with different vegetative compatibility genes undergo hyphal fusion, followed by cell death of the fused hyphal compartments. The fused hyphae between the three arrows are dead. [© H. L. Robinson & Jim Deacon, The University of Edinburgh]
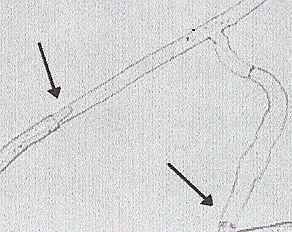
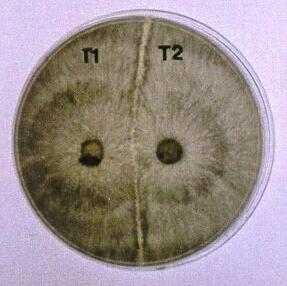
Fig. 9.7. Left: Part of Fig. 8.6c at higher magnification, showing regrowth of a hyphal tip into the dead hyphal compartment. Right: Vegetative incompatibility between two strains of Rhizoctonia solani (Basidiomycota) opposed on an agar plate. The clear demarcation zone between the colonies resulted from post-fusion death of the hyphae where the colony tips fused. [© P.M. McCabe, M.P.Gallagher & Jim Deacon, The University of Edinburgh]
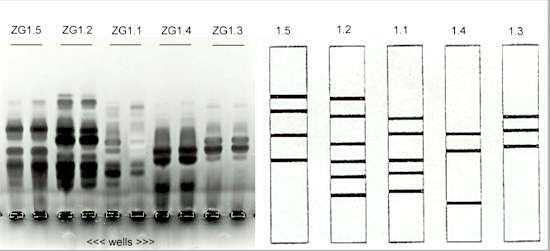
Fig. 9.8. Five distinctive zymogram groups (ZGs) of Rhizoctonia strains that cause bare patch disease of wheat in Australia (similar to crater disease, shown in Fig. 14.3). Protein extracts were run on acrylamide gels containing pectin then stained to develop the bands of pectic enzymes. [Courtesy of M. Sweetingham; from G.C.MacNish & M.W.Sweetingham (1993) Mycological Research 97, 1056-1058]
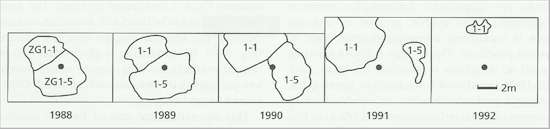
Fig. 9.9. Mapped positions of patches of stunted wheat plants caused by Rhizoctonia in a field trial site in Australia. Two adjacent patches were caused by different pectic zymogram groups (ZG1.1 and ZG1.5) which never invaded the territory of the other. The patches expanded and contracted in successive years. The central dot is a fixed reference point. [Based on G.C. MacNish et al. (1993) Australian Journal of Agricultural Research 44, 1161-1173]
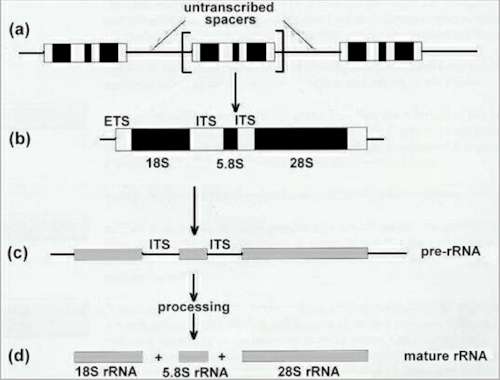
Fig. 9.10. Organisation and processing of the eukaryotic rRNA genes. |
|||||||||||